Indirect Reference Intervals
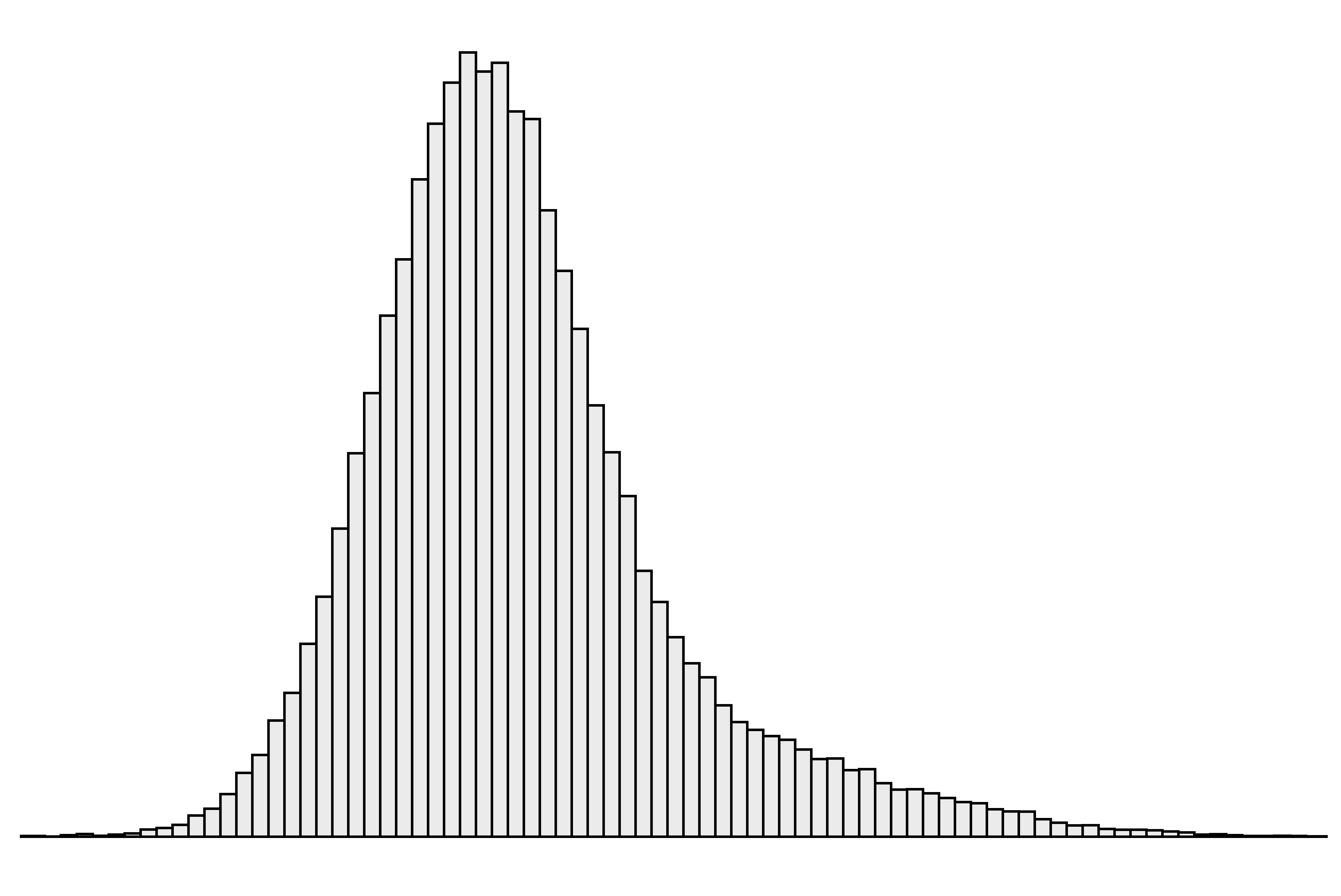
The ‘indirect’ approach to setting reference intervals uses results from samples collected for reasons other than a formal reference interval study.1 The results are generally extracted from pathology laboratory databases and therefore the data contains results both from individuals who are healthy and those with disease (Figure 1).
 |
|---|
| Figure 1. A simulated ‘mixed’ data set containing results both from individuals who are essentially healthy and those with disease. |
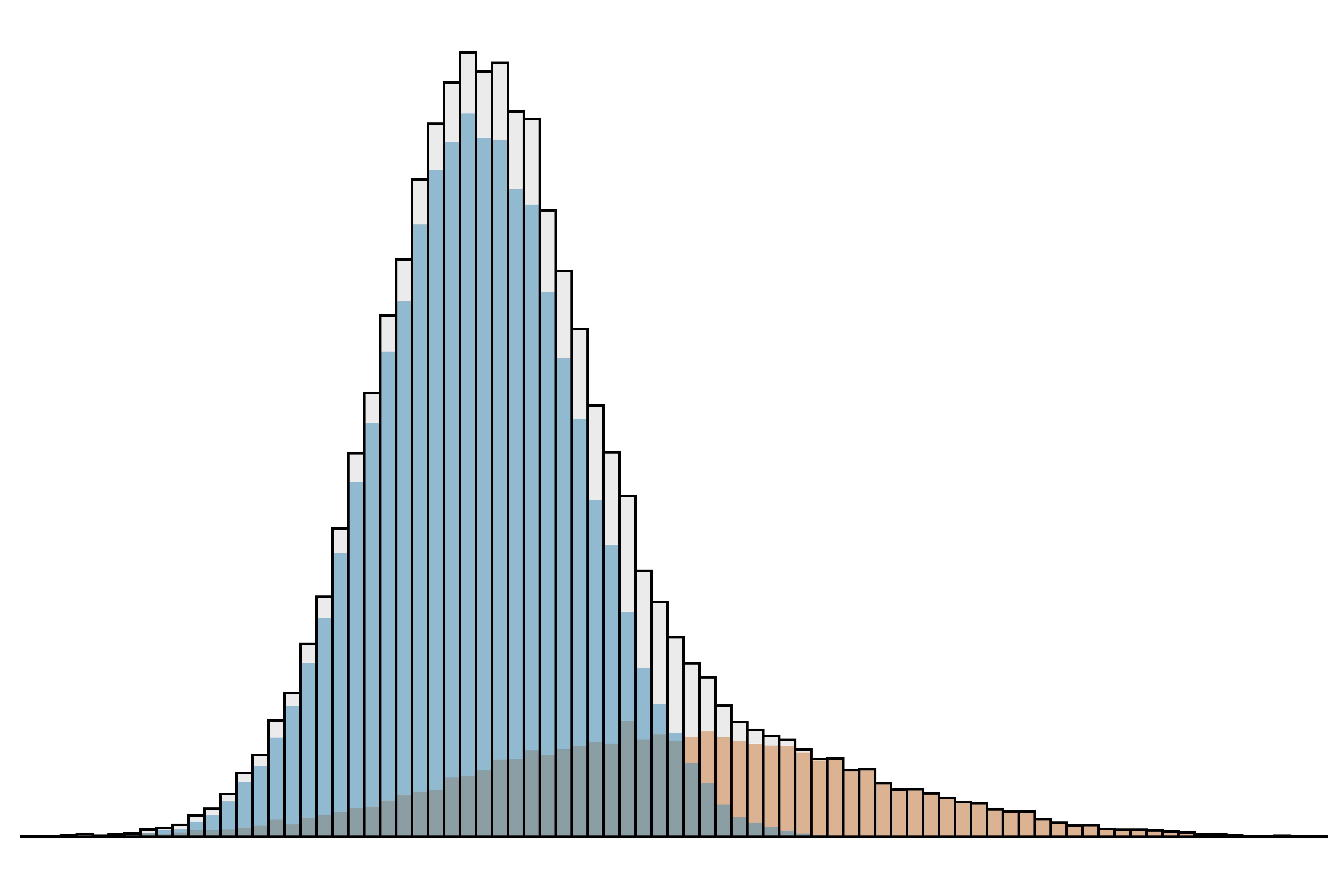
These two groups must somehow be separated. If it is possible to identify the patients with disease affecting the analyte of interest, these may be excluded and reference intervals set in the usual way. However, laboratories generally do not have access to detailed clinical information. The best approach then is try to identify the distribution of results from healthy individuals from among the ‘mixed’ data set (Figure 2). There are a number of techniques able to do this, including Bhattacharya analysis, the Hoffmann method and refineR. These techniques may be applied online using BhattApp, HoffApp and the RefineR App.
 |
|---|
| Figure 2. The mixed data set in Figure 1 can be resolved into two Gaussian distributions. |
There are some notable advantages of the indirect approach. It is faster and cheaper to perform than a formal reference interval study, its results reflect routine pre-analytical and laboratory conditions and, because it is easier to access large data sets, it can generate reference intervals for multiple age and sex partitions.
However, indirect approaches also are easier to get wrong than the direct approach; therefore they should be applied cautiously and their results critically evaluated. Nevertheless, the IFCC Committee on Reference Intervals and Decision Limits is supportive of indirect approaches and provides recommendations for their use.2
References
- Clinical and Laboratory Standards Institute. CLSI document EP28-A3c. Wayne, PA, USA: CLSI; 2010. Defining, Establishing, and Verifying Reference Intervals in the Clinical Laboratory; Approved Guideline – Third Edition.
- Jones GR, Haeckel R, Loh TP, et al. IFCC Committee on Reference Intervals and Decision Limits. Indirect methods for reference interval determination - review and recommendations. Clin Chem Lab Med 2018; 57: 20–9. PubMed